
In 2001, a group of Japanese scientists made a startling discovery at a rubbish dump. In trenches packed with dirt and waste, they found a slimy film of bacteria that had been happily chewing through plastic bottles, toys and other bric-a-brac. As it broke down the trash, the bacteria harvested the carbon in the plastic for energy, which it used to grow, move and divide into even more plastic-hungry bacteria. Even if not in quite the hand-to-mouth-to-stomach way we normally understand it, the bacteria was eating the plastic.
The scientists were led by Kohei Oda, a professor at the Kyoto Institute of Technology. His team was looking for substances that could soften synthetic fabrics, such as polyester, which is made from the same kind of plastic used in most beverage bottles. Oda is a microbiologist, and he believes that whatever scientific problem one faces, microbes have probably already worked out a solution. “I say to people, watch this part of nature very carefully. It often has very good ideas,” Oda told me recently.
What Oda and his colleagues found in that rubbish dump had never been seen before. They had hoped to discover some micro-organism that had evolved a simple way to attack the surface of plastic. But this bacteria was doing much more than that – it appeared to be breaking down plastic fully and processing it into basic nutrients. From our vantage point, hyperaware of the scale of plastic pollution, the potential of this discovery seems obvious. But back in 2001 – still three years before the term “microplastic” even came into use – it was “not considered a topic of great interest”, Oda said. The preliminary papers on the bacteria his team put together were never published.
In the years since the group’s discovery, plastic pollution has become impossible to ignore. Within that roughly 20-year span, we have generated 2.5bn tonnes of plastic waste and each year we produce about 380 million tonnes more, with that amount projected to triple again by 2060. A patch of plastic rubbish seven times the size of Great Britain sits in the middle of the Pacific Ocean, and plastic waste chokes beaches and overspills landfills across the world. At the miniature scale, microplastic and nanoplastic particles have been found in fruits and vegetables, having passed into them through the plants’ roots. And they have been found lodged in nearly every human organ – they can even pass from mother to child through breast milk.
Current methods of breaking down or recycling plastics are woefully inadequate. The vast majority of plastic recycling involvesa crushing and grinding stage, which frays and snaps the fibres that make up plastic, leaving them in a lower-quality state. While a glass or aluminium container can be melted down and reformed an unlimited number of times, the smooth plastic of a water bottle, say, degrades every time it is recycled. A recycled plastic bottle becomes a mottled bag, which becomes fibrous jacket insulation, which then becomes road filler, never to be recycled again. And that is the best case scenario. In reality, hardly any plastic – just 9% – ever enters a recycling plant. The sole permanent way we’ve found to dispose of plastic is incineration, which is the fate of nearly 70 million tonnes of plastic every year – but incineration drives the climate crisis by releasing the carbon in the plastic into the air, as well as any noxious chemicals it might be mixed with.
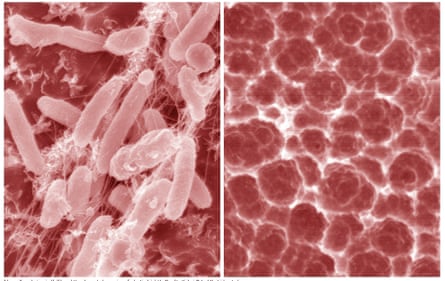
In the years after their discovery, Oda and his student Kazumi Hiraga, now a professor, continued corresponding and conducting experiments. When they finally published their work in the prestigious journal Science in 2016, it emerged into a world desperate for solutions to the plastic crisis, and it was a blockbuster hit. Oda and his colleagues named the bacteria that they had discovered in the rubbish dump Ideonella sakaiensis – after the city of Sakai, where it was found – and in the paper, they described a specific enzyme that the bacteria was producing which allowed it to break down polyethylene terephthalate (PET), the most common plastic found in clothing and packaging. The paper was reported widely in the press, and it currently has more than 1,000 scientific citations, placing it in the top 0.1% of all papers.
But the real hope is that this goes beyond a single species of bacteria that can eat a single kind of plastic. Over the past half-century, microbiology – the study of small organisms including bacteria and some fungi – has undergone a revolution that Jo Handelsman, former president of the American Society for Microbiology, and a science adviser to the Obama White House, described to me as possibly the most significant biological advance since Darwin’s discovery of evolution. We now know that micro-organisms constitute a vast, hidden world entwined with our own. We are only beginning to grasp their variety, and their often incredible powers. Many scientists have come around to Oda’s view – that for the host of seemingly intractable problems we are working on, microbes may have already begun to find a solution. All we need to do is look.
A discovery like Oda’s is only a starting point. To have any hope of mitigating this globe-spanning environmental disaster of our own making, the bacteria will have to work faster and better. When Oda and his group originally tested the bacteria in the lab, they placed it in a tube with a 2cm-long piece of plastic film weighing a 20th of a gram. Left at room temperature, it broke down the tiny bit of plastic into its precursor liquids in about seven weeks. This was very impressive and far too slow to have any meaningful impact on plastic waste at scale.
Fortunately, over the past four decades, scientists have become remarkably proficient at engineering and manipulating enzymes. When it comes to plastic chewing, “the Ideonella enzyme is actually very early in its evolutionary development”, says Andy Pickford, a professor of molecular biophysics at the University of Portsmouth. It is the goal of human scientists to take it the rest of the way.
When any living organism wishes to break down a larger compound – whether a string of DNA, or a complex sugar, or plastic – they turn to enzymes, tiny molecular machines within a cell, specialised for that task. Enzymes work by helping chemical reactions happen at a microscopic scale, sometimes forcing reactive atoms closer together to bind them, or twisting complex molecules at specific points to make them weaker and more likely to break apart.
If you want to improve natural enzyme performance, there are approaches that work in almost every case. Chemical reactions tend to work better at higher temperatures, for instance (this is why, if you want to make a cake, it is better to set the oven at 180C rather than 50C); but most enzymes are most stable at the ambient temperature of the organism they work in – 37C in the case of humans. By rewriting the DNA that codes an enzyme, scientists can tweak its structure and function, making it more stable at higher temperatures, say, which helps it work faster.
This power sounds godlike, but there are many limitations. “It is often two steps forward, one step back,” says Elizabeth Bell, a researcher at the US government’s National Renewable Energy Laboratory (NREL) in Colorado. Evolution itself involves tradeoffs, and while scientists understand how most enzymes work, it remains difficult to predict the tweaks that will make them work better. “Logical design tends not to work very well, so we have to take other approaches,” says Bell.

Bell’s own work – which focuses on PETase, the enzyme that Ideonella sakaiensis produces to break down PET plastics – takes a brute-force approach in order to turbocharge natural evolution. Bell takes the regions of the enzyme that work directly on plastic and uses genetic engineering to subject them to every possible mutation. In the wild, a mutation in an enzyme might occur only once in every few thousand times the bacteria divides. Bell ensures she gets hundreds, or thousands of potentially beneficial mutants to test. She then measures each one for its ability to degrade plastic. Any candidates that show even marginal improvement get another round of mutations. The head of the NREL research group, Gregg Beckham, refers to it as “evolving the crap out of an enzyme”. Last year, she published her latest findings, on a PETase enzyme she had engineered that could degrade PET many times faster than the original enzyme.
But building an enzyme that suits our purposes isn’t just a case of scientists tinkering until they get the perfect tool. Before the publication of Oda’s paper in 2016, no one knew that bacteria capable of digesting plastic existed. Now, we have one solidly documented case. Given that we have discovered only a tiny fraction of microbial life, a far better candidate might be out there. In engineering terms, we may currently be trying to squeeze elite racing performance out of a Toyota Yaris engine, when somewhere, yet to be discovered, there is the bacterial equivalent of a Ferrari. “This is something we constantly struggle with,” says Beckham. “Do we go back to the well to search and see if nature has the solution? Or do we take the small footholds we have to the lab and work on them now?”
This question has led to a boom in what is known as bioprospecting. Like panning for gold in a river, bioprospectors travel the world looking to discover interesting and potentially lucrative microbes. In 2019, a team at Gwangju National University in South Korea took a construction drilling rig to the municipal dump outside town, and drilled 15 metres under the trash trenches to reveal decades-old plastic garbage. In it, Prof Soo-Jin Yeom and her students found a variety of the bacteria Bacillus thuringiensis that appeared to be able to survive using polyethylene bags as food. Yeom’s team is now studying which enzymes the bacteria might be using, and whether it is really able to metabolise the plastic.
In vast mangrove swamps on the coastlines of Vietnam and Thailand, Simon Cragg, a microbiologist from the University of Portsmouth, is hunting for other potential PET-eating microbes. “The plastic-degrading enzymes we’ve already seen are quite similar to natural enzymes that degrade the coatings of plant leaves,” he told me. “Mangroves have a similar waterproof coating in their roots, and the swamps, sadly, also contain a shocking amount of plastic tangled up in them.” His hope is that a bacteria capable of degrading the mangrove roots will be able to make the jump to plastic.
For most of the roughly 200 years we have been seriously studying them, microbes were in a sort of scientific jail: mainly assumed to be pathogens in need of eradication, or simple workhorses for a few basic industrial processes, such as fermenting wine or cheese. “Even as recently as 40-50 years ago, microbiology was treated as a passe science,” Handelsman, the former American Society for Microbiology president, told me.
In the 20th century, as physics advanced to split the atom, and biologists came to classify many of the world’s plant and animal species, scientists who studied the very, very small domains of life lagged behind. But there were tantalising signs of the hidden world just beyond our reach. As early as the 1930s, microbiologists were puzzling over the disconnect between the microbial world they encountered in the wild and what they could study in the laboratory. They found that if they placed a sample – say a drop of seawater or smear of dirt – under a microscope, they could see hundreds of wondrous and varied organisms swirling about. But if they placed the same sample on to the gelatinous nutrient slurry of a petri dish, only a few distinct species would survive and grow. When they went to count the number of microbial colonies growing on the plate, it was a meagre handful compared to what they had just seen magnified. This would later be dubbed “the great plate count anomaly”. “With the microscope, and then the electron microscope, you could see all these hints. But these species wouldn’t grow on the plates, which is how we would characterise and study them,” said William Summers, a physician and historian of science at Yale.
Like a rare and exotic animal that cannot thrive in captivity, most micro-organisms didn’t seem suited for life in the lab. And so scientists were stuck with whatever could survive in their limited conditions. Yet there were some microbiologists who attempted to escape this straitjacket and discover the true extent of the microbial kingdom. The story of the discovery of penicillin by Alexander Fleming in 1928 is familiar: a fungal spore wafting through the corridors of St Mary’s hospital and settling at random in Fleming’s petri dish contained penicillin, which turned out to be one of the most potent medical weapons of the 20th century. Less well known, but no less significant, is the story of the Rutgers University chemist Selman Waksman, who coined the term “antibiotic” after noting that certain soil bacteria produced toxins that killed or inhibited other bacteria with whom they were competing for food. Waksman worked tirelessly to figure out the conditions required to grow these wild bacteria in his lab, and his efforts produced not just the second commercially available antibiotic, streptomycin, in 1946, but the next five antibiotics brought to market, too. Ultimately, searching the soil for antibiotic-producing microbes proved far more fruitful than waiting for them to float into one’s laboratory. Today, 90% of all antibiotics are descended from the grouping of bacteria that yielded Waksmans original discoveries.

Efforts like Waksman’s were relatively rare. It wasn’t until the discovery of simple chemical techniques to read the sequence of DNA – first emerging in the 1970s, but widely and commercially available from the mid-1980s – that things began to change. Suddenly the microbes under the microscope could be catalogued and identified by their DNA, which also hinted at how they might grow and function. Not only that, says Handelsman, “the genetic diversity we were seeing was enormous”. It turned out that “these life forms that looked quite similar are in fact very, very different. It opened this door to realising how much more was out there.”
About 25 years ago, the consensus among scientists was that there were probably fewer than ten million species of microbes on the planet; in the past decade, some new studies have put the number as high as a trillion, the vast majority still unknown. In our bodies, scientists have found microbes that affect everything from our ability to resist disease to our very moods. In the deep seas, scientists have found microbes that live on boiling thermal vents. In crude oil deposits, they have found microbes that have evolved to break down fossil fuels. The more we look, the more extraordinary discoveries we will make.
Their adaptability makes microbes the ideal companion for our turbulent times. Microbes evolve in ways and at speeds that would have shocked Darwin and his contemporaries. Partly because they divide quickly and can have population sizes in the billions, and partly because they often have access to evolutionary tricks unknown to more complex lifeforms – rapidly swapping DNA between individuals, for instance – they have found ways to thrive in extreme environments. And, at this historical moment, humans are creating more extreme environments across the globe at an alarming rate. Where other animals and plants have no hope of evolving a solution quickly enough to outpace their changing habitats, microbes are adapting fast. They bloom in acidified water, and are discovered chewing up some of the putrid chemicals we slough off into the natural world. Just as Kohei Oda suggested, for many of our self-created problems, they are proposing their own solutions.
Finding new microbes and tinkering with them in the lab are the first steps, but scientists know that the final leap – into what they tend to call “the real world” or “industry” – can be elusive. In the case of plastic-eating microbes, that leap has now been made. Since 2021, a French company named Carbios has been running an operation that uses a bacterial enzyme to process about 250kg of PET plastic waste every day, breaking it down into its precursor molecules, which can then be made directly into new plastic. It’s not quite composting it back into the earth itself, but Carbios has achieved the holy grail of plastic recycling, bringing it much closer to an infinitely recyclable material like glass or aluminium.
Carbios works out of a low-slung industrial facility in Clermont-Ferrand, on the very same grounds as the first Michelin tyre factory. But inside, it resembles less a noxious old factory and more an urban brewery, with processed plastic waste sitting inside great steel fermentation silos. There is the sound of liquid rushing through pipes, but no fumes or smell. Dirty plastic from recycling depots sits in bales, ready to be transformed.
The plastic is first shredded and then run through a machine that resembles an immense die-press, which freezes it and forces it through a tiny opening at great pressure. The plastic pops out as pellets – or nurdles, as they’re known – about the size of corn kernels. At the microscopic level, the plastic nurdle is much less dense than what plastic chemists call its original “crystalline” form. The fibres that make up the plastic were originally packed into a tight lattice that made them smooth and strong; now, while still intact, the fibres are further apart and slack, which gives the enzymes a bigger area to attack.
In the wild, the bacteria would produce a limited amount of plastic-targeting enzyme, and many other enzymes and waste products as well. To accelerate the process, Carbios pays a biotech company to harvest and concentrate huge amounts of pure plastic-digesting enzyme from bacteria. The Carbios scientists then place the plastic nurdles in a solution of water and enzyme, inside a sealed steel tank several metres high. In the adjoining lab where the process is tested, you can observe the reaction taking place in smaller vessels. Inside, the off-white plastic bits swirl about like the flakes in a snowglobe. As time goes on, the plastic erodes away, its components dissolving into the solution, leaving only a greyish liquid churning behind the glass. The liquid now contains not solid PET, but two liquid chemicals called ethylene glycol and terephthalic acid, which can be separated out and turned into new plastic.
The technique Carbios has developed appears to scale easily. Two years ago the company was recycling a few kilos of plastic in a lab; now it can do about 250kg a day. In 2025, it will open a much bigger facility near the border with Belgium, with the capacity to recycle more than 130 tonnes a day.

The reason France has a working plastic recycling factory that uses bacterial technology, but the US and China do not, is that the French state has made plastic waste an urgent priority, setting a target that by 2025 all plastic packaging used in France must be fully recycled. While environmental campaigners would prefer eliminating new plastic entirely, Macron is betting that some amount of high-quality new plastic will be needed in the coming decades, and has taken a personal interest in Carbios, singling them out for praise on his LinkedIn account. The pressure appears to be working. Some of France’s largest manufacturers – from L’Oréal to Nestlé, and the outdoor outfitter Salomon – have signed up with Carbios to take on their waste. As governments around the world begin the slow grind toward meeting their ambitious pledges to reduce plastic waste, more are likely to follow.
These factories aren’t a magic solution. The enzyme recycling process is a series of biological and chemical reactions, and as they scale up, you’re reminded that nature is a ruthless accountant. If you track the various inputs required, and the carbon emissions, you find that cleaning the plastic, then heating and freezing it, comes with a major energy cost. The chemical reaction itself turns the surrounding solution acidic, and so like an outdoor pool, chemical base must be constantly added to the solution to keep it close to neutral, which creates several kilograms of sodium sulphate as a byproduct each time the reaction runs. Sodium sulphate has many uses, including glassmaking and in detergents, but everything from manufacturing the chemical base, to moving the sodium sulphate on to further uses, adds environmental costs and logistical friction.
In a sunny conference room in the factory complex, Emmanuel Ladent, the Carbios CEO, told me that the company’s recycling process currently produces 51% fewer emissions than making new plastic (with the significant added benefits of no new oil drilled to make the plastic, and no net addition of plastics to the world). “Very good,” he concluded, “but the hope is we are just getting started.” Carbios has not released their analysis publicly, but several other scientists familiar with the field told me that halving emissions was within the best-case scenarios for this kind of recycling.
Carbios and the scientists behind it – the University of Toulouse biologists Alain Marty and Vincent Tournier – have been working in the field for more than a decade. While many other scientists began doing similar work after the publication of Oda’s discovery, Marty and Tournier started out in the mid-00s. They used a different enzyme, called leaf compost cutinase (LCC), which did not evolve to work on plastic, but which Marty and Tournier thought had the potential to do so. (The waxy coating of leaves, which the enzyme works on, bear a close similarity to plastic.) “It was a bit weak, and it didn’t work well with any kind of high temperature, but it was a good beginning,” Marty told me recently. Untold rounds of genetic engineering later, the enzyme clearly works.
Gregg Beckham of the NREL research group says that LCC is “a great enzyme, for sure. It takes names and kicks butt.” But he cautions that it is still imperfect. It prefers highly processed plastic, and it’s not good at working in the acidic soup its own reactions create. Beckham’s hope is that because the enzyme produced by Ideonella Sakeinsis probably evolved to specifically attack plastic, it will provide a better chassis to tinker with. There is, of course, an element of competition here, with scientists casting a sceptical eye over their rivals’ work. When I mentioned Beckham’s comment to Marty at Carbios, he replied: “Every time there’s a new enzyme discovered – most recently this Ideonella Sakiensis one – it creates a lot of buzz. And so we test them – they never work very well in our tests.” After almost 20 years of collaboration, he is loyal to his leaf compost cutinase.
Will highly evolved microbes really deliver us from the plastic crisis? Some scientists think the technology will remain limited. A recent critical review in the journal Nature noted that many kinds of plastics would probably never be efficiently enzymatically digested, because of the comparatively huge amount of energy required to break their chemical bonds. Andy Pickford, the professor at Portsmouth, is familiar with the limitations, but thinks many good targets still exist. “Nylon is tough but doable,” he says. “Polyurethanes, also doable.” The scientists at Carbios agree, predicting that they will have a process to recycle nylon within a few years. If those predictions come to pass, about a quarter of all plastics would become truly recyclable; if there turns out to be an enzyme match for all the plastics that are theoretically susceptible to being broken down, just under half of all plastic waste could be on the table.
Even so, what most scientists are working towards is a world in which enzymes are set to work turning old plastic into new plastic. This is frustratingly limited in scope. It makes economic sense – but it is still producing plastic, and using energy to do so. And while recycling may slow down new plastic production, it won’t help us claw back the unfathomable amount of plastic that we have already released into the world, much of which remains too widespread, difficult and dirty to recapture.
No one has yet found a microbe that can truly transform an untreated piece of plastic in the way they transform organic matter: starting with a pile of carbon – say, a human body – and leaving nothing but the indigestible skeletal bits within a year or so. When scientists find plastic-eating microbes on bottles at the dump, or on rafts of rubbish in the ocean, the best these microbes can do is a kind of light gnawing. Like a teething baby, they aren’t going to have much effect on anything that isn’t softened and spoon-fed to them.
But microbes do have the ability to nullify some of the planet’s most noxious toxins, cleansing entire landscapes in the process. This works best on chemicals that have been present on earth for millions of years, allowing microbes to develop a taste for them. When the Exxon Valdez dumped 41m litres of oil into the Gulf of Alaska in 1989, coverage of the cleanup focused on images of environmentalists scrubbing oil-sodden seals and puffins. But much of the actual oil removal was accomplished by bacteria that naturally feed on crude oil. Nearly 50,000kg of nitrogen fertiliser was spread along the shoreline to turbocharge bacterial growth. Similarly, when a former industrial site in Stratford, east London, was chosen for the 2012 Olympic Games, the committee charged with cleaning it up moved more than 2,000 dump trucks’ worth of soil contaminated with petroleum and other chemicals to sites where it was pumped full of nitrogen and oxygen for weeks, inducing a bloom of bacterial growth that consumed the toxins. The soil was returned to Stratford, and the Olympic park sits atop it now.
The question of whether the same could be accomplished with plastic in the environment has received far less interest – and funding – than the prospect of more effective recycling. “There is not exactly a market incentive to clean up our waste, whether it’s CO2, or plastic,” says Victor di Lorenzo, a scientist at the Spanish National Biotechnology Centre in Madrid, and an evangelist for the large-scale application of microbes to solve humanity’s problems. “There is a return on investment to recycle plastic. But who will pay for these larger-scale projects that would help wider society? This is something only public support would remedy.”
Aside from the market problem, there is also a legal one. Once a microbial species has been genetically engineered, almost every country restricts its release back into the wild without special permission – which is rarely granted. The reasons for this are obvious. In the 1971 science fiction story Mutant 59: The Plastic Eater, a virus with the ability to instantaneously melt plastic spreads across the world, knocking planes out of the air and collapsing houses. It is unlikely any plastic-eating bacteria would become that efficient, but perturbing microbes can have devastating consequences.
Di Lorenzo is convinced the danger of this kind of work is minimal. “With early GMOs, people turned on them. Scientists were arrogant. It seemed like it was all about dominating nature and making profits. But we have a chance to remake that conversation. We could enter a new partnership between science and the natural world. If we present it honestly to people, they can decide whether it’s worth the risk.”
The vision of a deeper partnership with microbes is a powerful one. The EU has funded several groups to develop microbes and enzymes to turn plastic into fully biodegradable materials, rather than just new plastic. Last year, a German group engineered the Ideonella sakaiensis PETase into a marine algae, noting that someday it could be used to break down microplastic in the ocean.
Oda is convinced we haven’t even scratched the surface. When he and his colleagues first found Ideonella at the dump nearly 20 years ago, it wasn’t working solo. “As soon as I saw the film of micro-organisms on the plastic, I knew it was many microbes working together,” Oda told me. His team realised that while Ideonella was breaking the plastic into its industrially valuable precursors, other microbes were stepping in to further chew those into simple nutrients the microbial community could use. They were symbiotic. Partners, in a way. Oda has since written several papers pointing out that microbial communities might be developed into a system to remove micro- and nanoplastics from the soil. But he has received little interest.
In our conversations, Oda repeatedly bemoaned the lack of truly world-changing ideas coming from people who wanted to commercialise the discoveries he and his colleagues had made. There was an incredible amount of excitement about a factory that could turn old plastic into new; far less, it seemed, about one that could turn plastic back into water and air.
